Protein Expression Optimization Services
AIse Bio's Alchemist™ applies deep learning to predict and optimize protein expression in E. coli, S. cerevisiae, and CHO cells. Reduce wet-lab iterations by prioritizing high-expressing protein variants and designing mutations that maintain structure and activity.
Expression Ranking
Submit protein sequences to receive an AI-ranked list based on predicted expression levels. Focus resources on top candidates for E. coli, S. cerevisiae, or CHO expression.
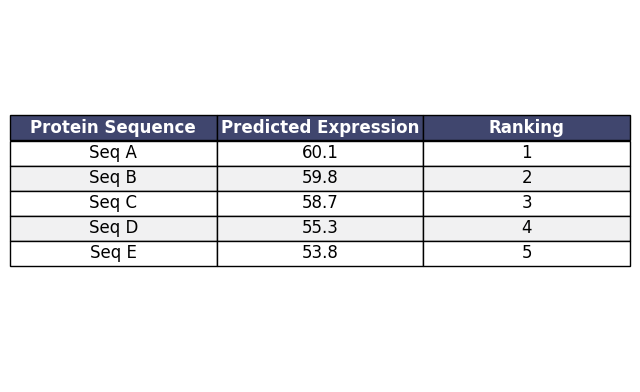
Expression Optimization
Generate AI-designed mutations that increase expression while conserving essential functional domains. Export prioritized variants for rapid experimental validation.
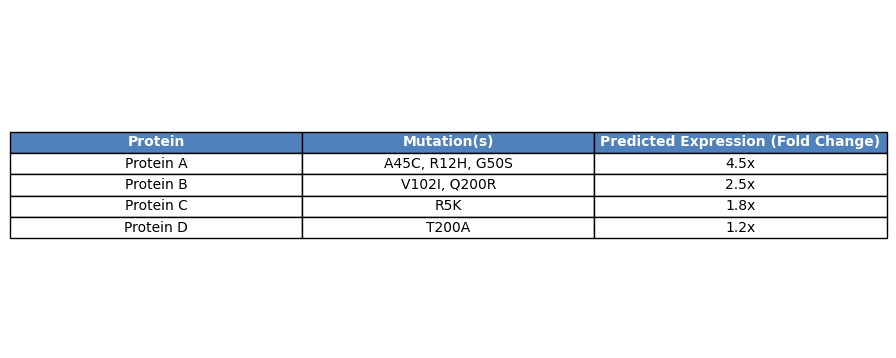
Supported Hosts
Alchemist™ supports E. coli, S. cerevisiae, and CHO cells, with additional organism-specific models in development.
Why Choose Aise Bio?
Experience the benefits of our AI-driven approach to protein expression optimization.
Faster Results
Accelerate your project and skip months of trial-and-error. Get AI-optimized protein expression predictions in days.
Cost Savings
Reduce the cost of your wet lab experiments by 90% or more and streamline your R&D budget.
Scientific Precision
Powered by deep learning, our models predict expression levels with high accuracy.
Data Security
We guarantee full client confidentiality and you own any IP that you create.
World-Class Expertise
Developed by top experts in protein engineering and AI/ML.
Our Technology
Our best-in-class AI deep learning and sequence-structure modeling technologies power our predictions. Our expression ranking models have been benchmarked and outperform the best published methods for E. coli and S. cerevisiae and we have many more organism specific models currently in development. Our mutation generation methods ensure that protein variants are highly likely to express well in your chosen host while preserving essential structure and function.
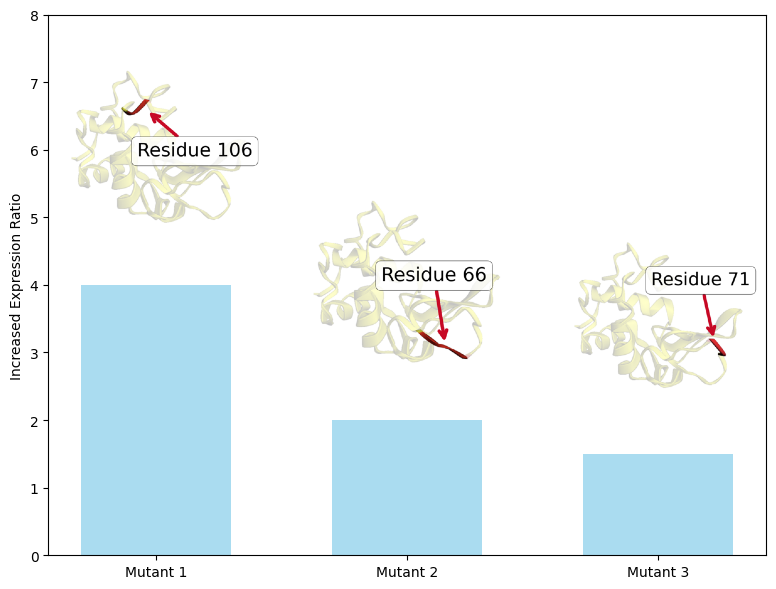
Get Started
Send your sequence or dataset and receive ranked predictions and optimized variants within days.